The LuminoDx miRapid® miRNA detection assay offers a single setup, single-plex or multiplex RT-qPCR solution for analyzing miRNAs. miRNA were reverse transcribed, followed by annealing and template extension, then real-time PCR and detection with Taqman probes. The high accuracy reagent system delivers consistent, sensitive and reproducible results.
Main Advantages:
1. One setup reaction set up: miRNA RT-PCR reaction can be set up in minutes, with the entire process conducted in a single tube with one setup only.
2. Efficient Process: The complete reaction takes approximately 2 hours.
3. Multiplex Detection: High possibility of multiplexing capability for miRNA assay development which is idea solution for limit sample.
4. Readiness: Multiple housekeeping miRNA assays have been developed, with customized development available upon request.
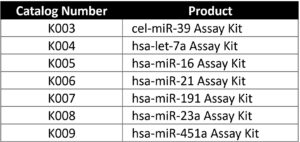
The reagent list below includes all of the necessary reagents to carry out accurate qRT-PCR detection of miRNA. The kit components consist of three elements: an enzyme mix, a reaction buffer, and an oligo mix (primers, probes, and linkers) for the miRNA of interest.
We can develop customized miRNA assay to serve your specific need. The typical development time is 4-8 weeks depending on the target and specific requirements. We run aggressive validation to ensure a robust and reproducible assay for your miRNA research.
LuminoDx has extensive expertise in isolating and analyzing miRNAs. We develop customized assays tailored to your specific miRNAs of interest and provide miRNA isolation and detection service. We warranty the quality of our work and commit a fast turnaround time to expedite your service need.

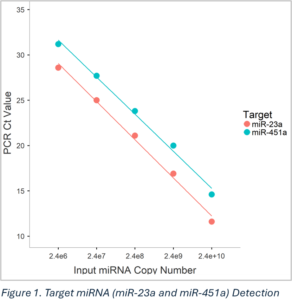
Detection Efficiency
To assess RT-PCR efficiency, mimic miR-23a and miR-451a were pooled and serially diluted as inputs for the PCR reaction.
miR-23a: Correlation coefficient = -0.997
miR-451a: Correlation coefficient = -0.996
The limit of detection (LOD) for both targets is comparable to ThermoFisher’s TaqMan Advanced miRNA assay.
Detection Reproducibility and Normalization
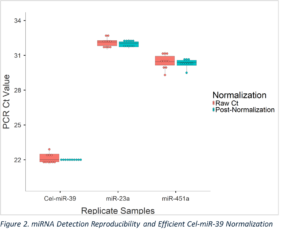
RNA was isolated from 10 replicates of normal human serum using the Thermo Fisher MagMAX mirVana total RNA isolation kit.
Cel-miR-39 was spiked into the samples during isolation, and RNA was eluted in 50 µL of resuspension buffer. RT-PCR was performed on the isolated RNA samples using the miRapid-PCR miRNA Assay – Panel A.
Normalization was achieved by adjusting all samples to Cel-miR-39 median Ct value of 22.0. Specifically, miR-23a and miR-451a Ct values were corrected for each sample based on the difference between their Cel-miR-39 Ct values and 22.0. This normalization resulted in more consistent detection of miR-23a and miR-451a, with a reduced range (miR-23a: from 1.2 to 0.8; miR-451a: 2.0 to 1.2) to and lower coefficients of variation (miR-23a: from 1.2% to 0.8%; miR-451a: 2.0% to 1.2%).
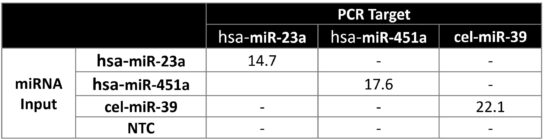
Specificity and off-Target Detection
No non-specific amplification was observed for any of the three target miRNA genes from normal human serum samples shown in Figure 2.